FastSurfer
After you run the recon-all with a tutorial data, I bet you might wonder how come I can skip this process time. The first solution would be FastSurfer.
FastSurfer is a fast and deep-learning pipeline for the fully automated processing of structural human brain MRIs. It provides conform outputs$
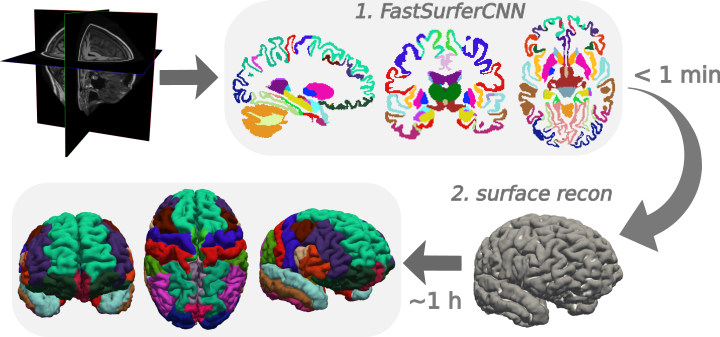
FastSurfer consists of two main parts:
FastSurferCNN Volumetric Segmentation.
FastSurferCNN is an advanced deep learning pipline for whole brain segmentation into 95 classes in under 1 minute, mimicking FreeSurfer’s anatomical segmentation and cortical parcellation.
go to Here either use git clone from you home directory to ge the file or download the file and put it in your home directory.
Set up and run FastSurfer
Set the path export FREESURFER_HOME=/usr(usrname)/local/freesurfer/6.0.0
source $FREESURFER_HOME/SetUpFreeSurfer.sh to activate the Freesurfer
datadir=/home/user/mri_data_directory
fastsurferdir=/home/user/fastsurfer_analysis_directory
Run FastSurfer:
./run_fastsurfer.sh --t1 $datadir/subject1/orig.mgz \
--sid subject1 --sd $fastsurferdir \
--parallel --threads 4 --py $(which python)(optional)
--sd Output directory $SUBJECTS_DIR
--sid Subject ID for directory inside $SUBJECTS_DIR to be created
--t1 T1 full head input. The network was trained with confirmed images, these specifications are checked in the eval.py script and the image is automatically conformed if it does not comply.
parallel means we use parallel computing to run 4 threads indicated by threads 4
--py $(which python) specify the python Fastsurfer use
Before you run the script, just ensure you check all the required packages
sed -i "s/==/>=/g" requirements.txt and pip install --no-index -r requirements.txt might help
It is worth to notice that FastSufer needs the support from FreeSurfer and it is works for FreeSurfer6.0.0 now